Taking a data-driven approach to precision medicine
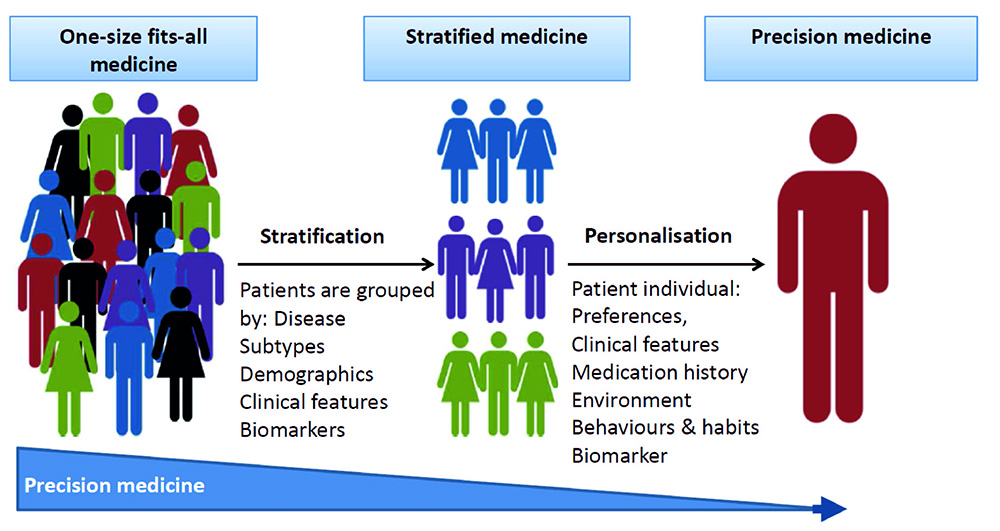
Precision medicine, or personalized medicine, combines data science tools with medical knowledge to enable physicians to more accurately predict disease susceptibility or prognosis, and develop a tailor-made treatment or prevention plan. In other words, the goal is to provide the right treatment to the right patient at the right time.
Data science research using electronic medical records has accelerated progress in the field of precision medicine. Kai Yang, professor of industrial and systems engineering, and Ph.D. student Milad Zafar Nezhad lead an interdisciplinary team of Wayne State researchers developing two novel predictive and feature learning approaches to precision medicine.
A deep learning method termed SAFS (Stacked Auto-encoders Feature Selection) presents a fresh approach to disease risk factor selection and prediction.
"Feature selection means 'important variable selection' or important factor selection.' It allows us to select the most critical factors-such as BMI, weight, blood sugar-for certain diseases,'" said Yang.
SAFS employs a powerful, unintuitive, and indirect methodology based on neural networks for feature representation, or converting raw data to features, because many raw data are not directly meaningful factors. One study focused on African-Americans at high risk for hypertension and other cardiovascular complications. The SAFS model was applied to find risk factors that affected left ventricular mass indexed to body surface area (LVMI), a major indicator of cardiovascular disease. Analysis of data obtained from more than 700 patients at Detroit Receiving Hospital showed that SAFS outperformed other popular methods in term of predicting LVMI and discovering critical risk factors of the disease.
The team also proposed a survival analysis framework using deep learning and active learning called Deep Active Survival Analysis (DASA). The motivation for this study comes from either literature gaps or application needs in several healthcare domains where the labeled data is scarce and high-dimensional.
"Survival analysis is a kind of statistical modeling where the main goal is to analyze and model time until the occurrence of an event of interest, such as patient death," said Nezhad. "DASA is able to improve the survival analysis performance significantly for risk prediction and survival time estimation, and provide treatment recommendations."
In this study, researchers used SEER-Medicare prostate cancer data to evaluate the performance of their approach and provide specific racial therapy insights based on different treatment plans among African-American and white patients.
SAFS and DASA each offer unique characteristics to further precision medicine and the customization of health care. Among Yang and Nezhad's collaborators for these studies were Dongxiao Zhu, associate professor of computer science; Jennifer Beebe-Dimmer, associate professor in the WSU School of Medicine and the Karmanos Cancer Institute; Julie Ruterbusch, a research assistant at Karmanos and the School of Medicine; and Phillip Levy, M.D., M.P.H., the Edward S. Thomas Endowed Professor of Emergency Medicine and assistant vice president of Translational Science and Clinical Research Innovation for WSU.